
Lennart Nilsson, Professor
![]()
Theoretical and simulation methods give a very detailed description
of biomolecular systems which can serve, not only to fill in the gaps between
the experiments, but also to provide predictions and suggestions for further
studies. By combining molecular dynamics simulations on a series of related
systems with experimental studies of basic physical/chemical properties
and structural studies new insights in the underlying mechanisms and processes
can be obtained. Since most biomolecular reactions take place in an aqueous
environment it is also very important to consider the effects of the surroundings,
which may be of several kinds: competition for hydrogen bonds, screening
of electrostatic interactions or favoring structural arrangements with
minimal exposure of hydrophobic groups, to name but a few possibilities.
Current projects include studies of stacking-unstacking of nucleotides,
binding of the glucocorticoid receptor to DNA, helix forming ability of
short peptides in mixed solvents and model building of various proteins.
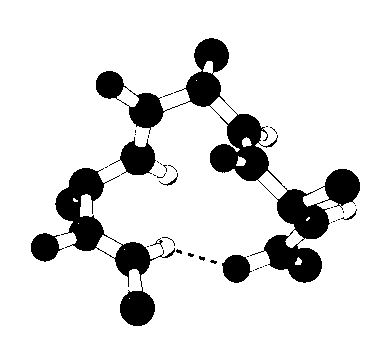
One turn of an alpha-helix used in study of helical stability (Brooks&Nilsson,
1993)
![]()
Recent departures
Pekka Mark,
PhD, TU Munich
Nicolas Foloppe, Ph.D., Ribotargets, Cambridge, UK
Mats Eriksson, Ph.D.
Srikanta Sen, Ph.D., Indian Institute of Chemical Biology, Calcutta
Ansuman Lahiri, PhD
Valere Lounnas, Ph.D
Yun Tang,
Ph.D
Joanna Sarzynska,
Ph.D., Institute of Bioorganic Chemistry, Polish Academy of Sciences
![]()
Vogel, H., Nilsson, L., Rigler, R., Meder, S.,Boheim, G., Beck, W., Kurth, H.H. & Jung, G. (1993) Structural Fluctuations Between Two Conformational States of a Transmembrane Helical Peptide Are Related to Its Channel Forming Properties in Planar Lipid Membranes, Eur. J. Bioch. 212, 305-313.
Whitley, P., Nilsson, L. & von Heijne, G. (1993) A Model for the 3D-Structure of the Membrane Domain of Escherichia coli Leader Peptidase Based on Disulphide Mapping, Biochemistry 32, 8534-8539.
Brooks, C.L. III & Nilsson, L (1993) The Promotion of Helix Formation in Peptides Dissolved in Alcohol and Water-Alcohol Mixtures, J. Am. Chem. Soc., 115, 11034-11035
Vihinen, M., Nilsson, L. & Smith, C.I.E. (1994) Tec homology (TH) adjacent to the PH domain, FEBS Letters 350, 263-265.
Vihinen, M., Zvelebil, M.J.J.M., Zhu, Q., Brooimans, R.A., Ochs, H.D., Zegers, B.J.M., Nilsson, L., Waterfield, M.D. & Smith, C.I.E. (1995) Structural Basis for Pleckstrin Homology Domain Mutations in X-linked Agammaglobulinemia, Biochemistry 34, 1475-1481.
Zilliacus, J., Wright, A.P.H., Carlstedt-Duke, J., Nilsson, L. & Gustafsson, J.-Å. (1995) Modulation of DNA Binding Specificity Within the Nuclear Receptor Family by Substitutions at a Single Amino Acid Position, PROTEINS 21, 57-67.
Eriksson, M., Berglund, H., Härd, T. & Nilsson, L. (1993) A Comparison of 15N NMR Relaxation Measurements with a Molecular Dynamics Simulation: Backbone Dynamics of the Glucocorticoid Receptor DNA-binding Domain, PROTEINS, 17, 375-390.
Eriksson, M.A.L., Härd, T. & Nilsson, L. (1995) Molecular Dynamics Simulations of the DNA -Binding Domain of the Glucocorticoid Receptor as a Dimer in Complex with DNA and as a Monomer in Solution, Biophysical Journal 68, 402-426.
Eriksson, M.A.L. & Nilsson, L. (1995) Structure, thermodynamics and cooperativity of the glucocorticoid receptor DNA-binding domain in complex with different response elements. Molecular dynamics simulation and free energy perturbation studies, J. Mol. Biol. 253, 453-472.
Nilsson, L. (1998) Protein Nucleic Acid Interactions. In "Encyclopedia of Computational Chemistry" (Eds. P. von Ragué Schleyer, N.L. Allinger, T. Clark, J. Gasteiger, P.A. Kollman, H. F. Schaefer III) , Vol. 3, 2220-2229, John Wiley & Sons.
Eriksson, M.A.L. & Nilsson, L. (1999) Structural and Dynamic Differences of the Estrogen Receptor DNA-binding Domain, Binding as a Dimer and as a Monomer to DNA. Molecular Dynamics Simulations Studies, European Biophysics Journal 28, 102-111.
Tang, Y. & Nilsson, L. (1999) Effect of G40R Mutation on the Binding of Human SRY Protein to DNA: A Molecular Dynamics View, PROTEINS 35, 101-113.
Tang, Y. & Nilsson, L. (1999) Molecular Dynamics Simulation of the Complex between Human U1A Protein and Hairpin II of U1 snRNAand of the Free RNA in Solution, Biophysical Journal, in press.
Norberg, J. & Nilsson, L. (1994) Stacking-Unstacking of the Dinucleoside Monophosphate Guanylyl-3'-5'-Uridine Studied with Molecular Dynamics, Biophysical Journal 67, 812-824.
Norberg, J. & Nilsson, L. (1995) Stacking Free Energy Profiles for All 16 Natural Ribodinucleoside Monophosphates in Aqueous Solution, J. Am. Chem. Soc., 117,10832-10840.
Norberg, J. & Nilsson, L. (1995) NMR Relaxation Times, Dynamics and Hydration of a Nucleic Acid Fragment from Molecular Dynamics Simulations, J. Phys. Chem. 99,14876-14884.
Norberg, J. & Nilsson, L. (1995) Temperature Dependence of the Stacking Propensity of Adenylyl-3',5'-Adenosine, J. Phys. Chem. 99, 3056-3058.
Norberg, J. & Nilsson, L. (1995) Potential of Mean Force Calculations of the Stacking-Unstacking Process in Single-stranded Deoxyribodinucleoside Monophosphates, Biophysical Journal 69, 2277-2285..
Sen, S. & Nilsson, L. (1998) Molecular Dynamics of Duplex
Systems Involving PNA: Structural and Dynamical Consequences of the Nucleic
Acid Backbone, J. Am. Chem. Soc. 120, 619-631