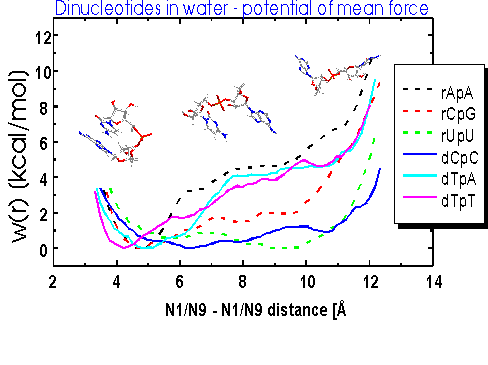
The phenomenon of base stacking, which we treat here in full atomic detail for RNA fragments including solvent and the sugar phosphate backbone, is not yet completely understood. We present free energy profiles of stacking for all 16 natural ribodinucleoside monophosphates based on potential of mean force calculations for this unimolecular process. Stacking preferences follow the general sequence purine-purine > purine-pyrimidine pyrimidine-purine > pyrimidine-pyrimidine with the stacked state having 2-6 kcal/mol lower free energy than the unstacked states for purine-purine combinations. The ribodinucleoside monophosphates containing both a purine and a pyrimidine base showed in most cases higher stacking propensities if the purine base was in the 5'-position. No or a very small free energy barrier was observed for the unstacking of the pyrimidine-pyrimidine ribodinucleoside monophosphates. A large number of different structures, both stacked and unstacked, were obtained and major transitions of the backbone torsion angles were observed. The pseudorotation phase angle showed a preference for the 2E mode in the unstacked states of ribodinucleoside monophosphates containing pyrimidine-pyrimidine combinations. The strongest interactions with solvent were observed for the phosphate oxygens, and the direct interactions of the 2'OH group of the 5'-ribose with the 3' base and ribose destabilize the stacked state by about 2 kcal/mol. Both the solvent and the sugar-phosphate backbone influence the stacking preferences of the dimers.
